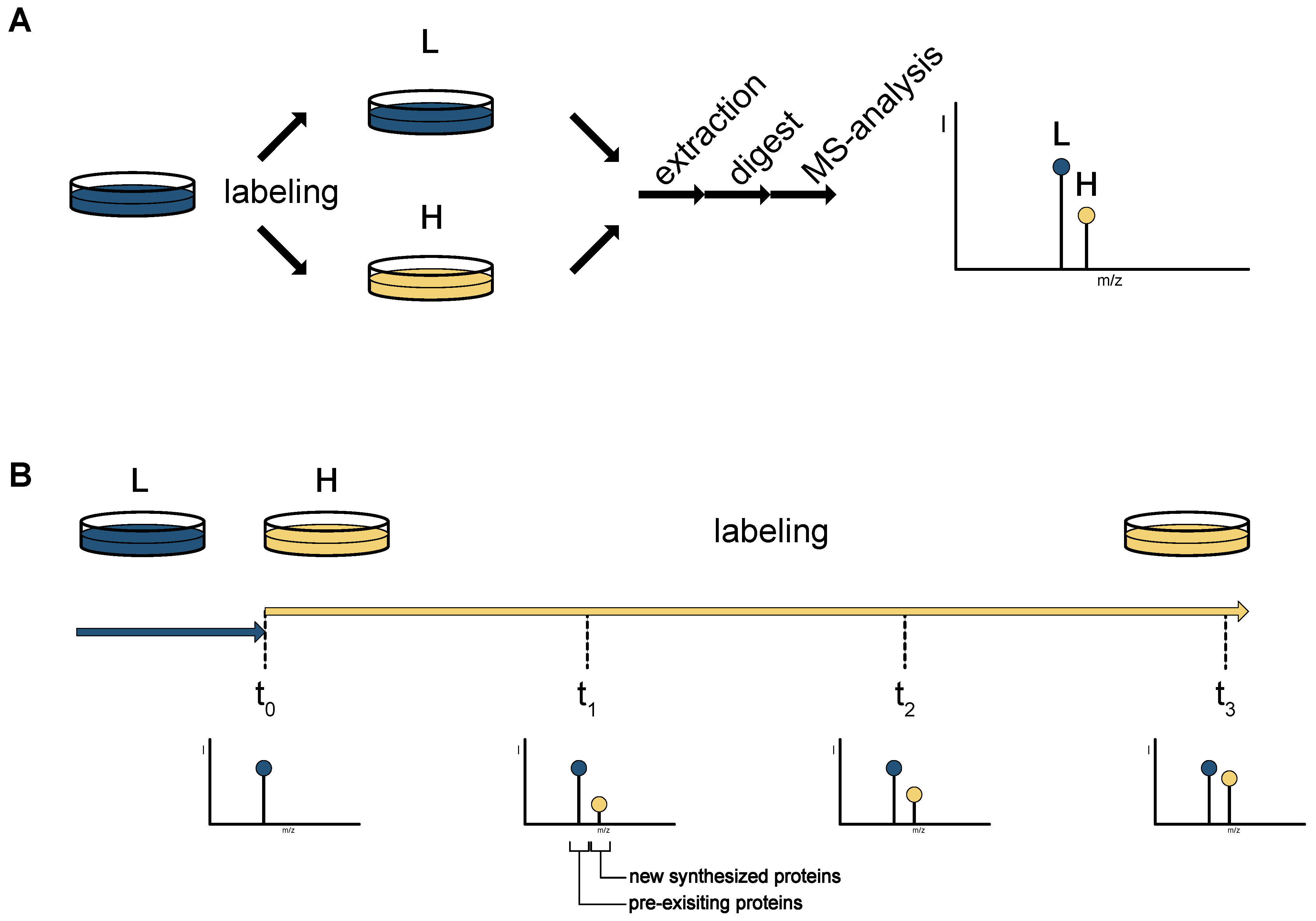
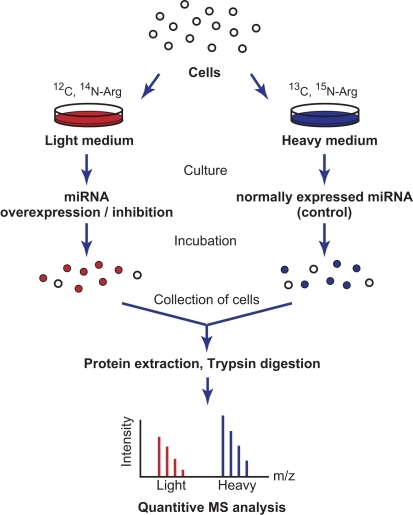
PLOS ONE: Plant SILAC: Stable-Isotope Labelling with Amino Acids of Arabidopsis Seedlings for Quantitative Proteomics

Cells can be metabolically labeled with a combination of Aha and/or... | Download Scientific Diagram

Light or heavy isotope labeled amino acids (Lys, +6 Da; Arg, +10 Da)... | Download Scientific Diagram
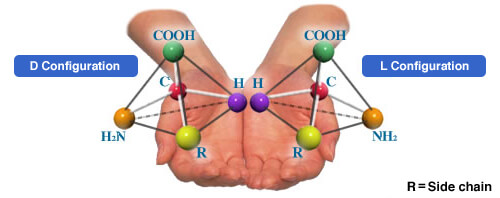
What are L-, D-, and DL-Amino Acids? | Enhancing Life with Amino Acids | About us | Ajinomoto Group Global Website - Eat Well, Live Well.

Schematic representation of a stable isotope labelling with amino acids... | Download Scientific Diagram

Quantitative proteomics - SILAC Harini Chandra a The identification and quantitation of complex protein mixtures have been facilitated by mass spectrometric. - ppt download

Amino acid based labeling techniques. (A) SILAC labeling. Proteins are... | Download Scientific Diagram

Characterization of the Tyrosine Kinase-Regulated Proteome in Breast Cancer by Combined use of RNA interference (RNAi) and Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) Quantitative Proteomics* - Molecular &

Figure 3 | Proteomics progresses in microbial physiology and clinical antimicrobial therapy | SpringerLink

Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics | Nature Protocols
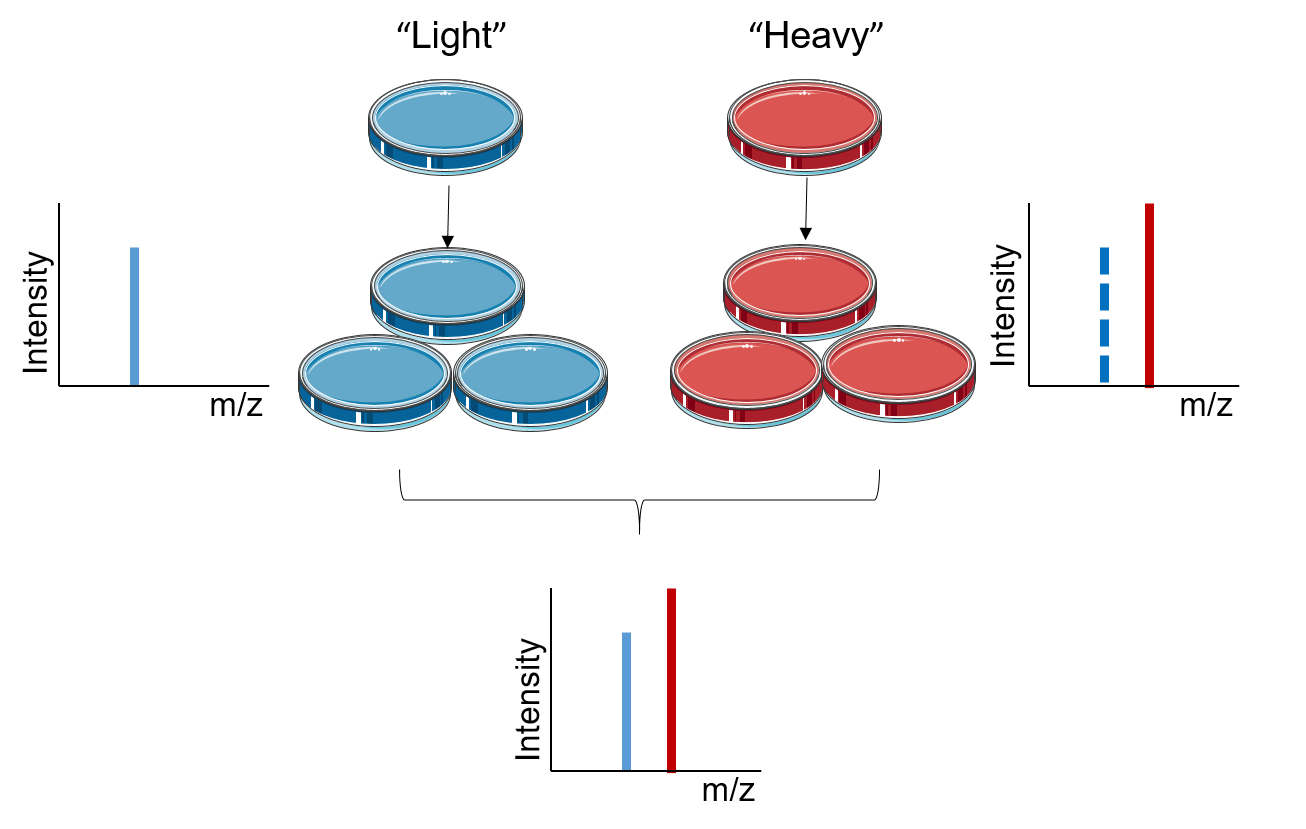
Stable Isotope Labeling Using Amino Acids in Cell Culture (SILAC): Principles, Workflow, and Applications – Creative Proteomics Blog

IJMS | Free Full-Text | SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers | HTML